Getting Started with GPS Data Analysis in Python
Possibly the last thing the world needs is another gps analysis tool, but I have been recording my bike rides, hikes, walks, runs, and even canoe and 4x4 trips consistently since 2015, so it seems like a good data set to work from to get beyond the basics in Python, data conditioning, analysis, and visualization.
Understanding the Data Source
Whether recorded using your phone, sports watch, bike computer, or handheld, most services will output GPS data in the form of a .gpx file. I use a web service called Tapiriik to export all activities I upload to RideWithGPS, Strava, or Garmin Connect to a folder in Dropbox, so I’ve already got a complete set of .gpx files to work with.
Here’s a sample of the .gpx file we’ll be working from:
<?xml version='1.0' encoding='UTF-8'?>
<gpx xmlns:gpxtpx="http://www.garmin.com/xmlschemas/TrackPointExtension/v1" xmlns:gpxext="http://www.garmin.com/xmlschemas/GpxExtensions/v3" xmlns:gpxdata="http://www.cluetrust.com/XML/GPXDATA/1/0" xmlns="http://www.topografix.com/GPX/1/1" creator="tapiriik-sync">
<metadata>
<name>Midnight Massacre singlespeed </name>
</metadata>
<trk>
<name>Midnight Massacre singlespeed </name>
<trkseg>
<trkpt lat="33.48812802" lon="-97.16527113">
<time>2017-08-05T23:59:57+00:00</time>
<ele>220.0</ele>
</trkpt>
<trkpt lat="33.48812882" lon="-97.16526836">
<time>2017-08-05T23:59:59+00:00</time>
<ele>220.0</ele>
</trkpt>
...
<trkpt lat="33.48796018" lon="-97.16536649">
<time>2017-08-06T02:17:50+00:00</time>
<ele>219.8</ele>
</trkpt>
<trkpt lat="33.48797073" lon="-97.16536446">
<time>2017-08-06T02:17:52+00:00</time>
<ele>219.8</ele>
</trkpt>
</trkseg>
</trk>
</gpx>
You can see it’s just an XML document with a standardized format for representing waypoints, tracks, and routes. The basic component of the GPX file is the <trkpt> or track-point. At minimum, the <trkpt> includes longitude, latitude, and a timestamp. It may also include elevation, either from a barometer on the device, or added in post-processing from a digital elevation model. My Garmin Vivoactive 3 also includes heart rate from the onboard optical heart rate sensor, and cadence from an additional ant+ sensor on the crank-arm of my bike. These attributes are stored in a sub-level of the <trkpt> called <extensions>.
<trkpt lat="32.845418" lon="-96.704896">
<time>2020-07-27T13:17:37+00:00</time>
<ele>145.0</ele>
<extensions>
<gpxtpx:TrackPointExtension>
<gpxtpx:hr>75</gpxtpx:hr>
<gpxtpx:cad>0</gpxtpx:cad>
</gpxtpx:TrackPointExtension>
</extensions>
</trkpt>
For this exercise, we’ll stick to an older file recorded with my phone and won’t be dealing with the extra attributes yet.
I followed along with these articles before doing my own thing here:
- How tracking apps analyse your GPS data
- Analysis and visualization of activities from Garmin Connect
Loading the data
Since GPX files are just XML, we could write a script to parse the records ourselves, but for this we’ll be using the gpxpy library. gpxpy creates a “gpx object”, a nested set of arrays for tracks, segments, and points, respectively. The points array contains the data from <trkpt> as attributes: longitude, latitude, elevation, timestamp, and extension. points.extension contains the additional attributes and is accessed with:
ext = point.extensions[0].getchildren()[0]
hr = int(ext.text)
We’ll pour the data from the gpx object into a Pandas dataframe object, a data table or lightweight database held in memory with lots of useful high-level functions for data manipulation and analysis.
import gpxpy
import pandas as pd
gpx_file = open('2017-08-05_Midnight Massacre singlespeed _Cycling.gpx', 'r') # open the file in read mode
gpx_data = gpxpy.parse(gpx_file)
Let’s have a look at the gpx object gpx_data and see how data is organized within it.
len_tracks = len(gpx_data.tracks)
len_segments = len(gpx_data.tracks[0].segments)
len_points = len(gpx_data.tracks[0].segments[0].points)
print(f"Tracks: {len_tracks} \nSegments: {len_segments} \nPoints: {len_points} \n")
Tracks: 1
Segments: 1
Points: 3774
Looks like there is only 1 Track with 1 Segment with 3774 Points.
Since the point data is burried under an unweildy path, lets make a variable as a shortcut to it. Then we’ll iterate through the points and append them to a dataframe.
gpx_points = gpx_data.tracks[0].segments[0].points
df = pd.DataFrame(columns=['lon', 'lat', 'elev', 'time']) # create a new Pandas dataframe object with give column names
# loop through the points and append their attributes to the dataframe
for point in gpx_points:
df = df.append({'lon' : point.longitude, 'lat' : point.latitude, 'elev' : point.elevation, 'time' : point.time}, ignore_index=True)
Exploratory Data Analysis & Cleanup
Now that that the data is in a dataframe, it’s ready for some basic Exploratory Data Analysis to get a feel for the data set, how it’s organized, and see if any cleanup is needed. Since the data came from a well-formatted XML file, we know its structure, but for the sake of practice lets run through a few things anyway.
I’ll start with a look at the first n rows of the data with df.head(n) and a couple of quick summaries of the dataframe with df.info() and df.describe().
print(f"df.head(10) displays the first 10 rows of the dataframe:\n\n{df.head(10)}\n\n")
print(f"df.info() gives a summary of the dataframe's contents and data-types:\n")
print(df.info(), "\n\n") # I had to pull this out separately because it was outputting the info table before the text, no idea why.
print(f"df.describe() gives a basic statistical summary of the numerical data:\n\n{df.describe()}")
df.head(10) displays the first 10 rows of the dataframe:
lon lat elev time
0 -97.165271 33.488128 220.0 2017-08-05 23:59:57+00:00
1 -97.165268 33.488129 220.0 2017-08-05 23:59:59+00:00
2 -97.165268 33.488129 220.0 2017-08-06 00:00:01+00:00
3 -97.165269 33.488129 220.0 2017-08-06 00:00:03+00:00
4 -97.165269 33.488130 220.0 2017-08-06 00:00:05+00:00
5 -97.165269 33.488129 220.0 2017-08-06 00:00:07+00:00
6 -97.165268 33.488130 220.0 2017-08-06 00:00:09+00:00
7 -97.165267 33.488127 220.0 2017-08-06 00:00:11+00:00
8 -97.165264 33.488127 220.0 2017-08-06 00:00:13+00:00
9 -97.165261 33.488125 220.0 2017-08-06 00:00:15+00:00
df.info() gives a summary of the dataframe's contents and data-types:
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 3774 entries, 0 to 3773
Data columns (total 4 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 lon 3774 non-null float64
1 lat 3774 non-null float64
2 elev 3774 non-null float64
3 time 3774 non-null datetime64[ns, SimpleTZ("Z")]
dtypes: datetime64[ns, SimpleTZ("Z")](1), float64(3)
memory usage: 118.0 KB
None
df.describe() gives a basic statistical summary of the numerical data:
lon lat elev
count 3774.000000 3774.000000 3774.000000
mean -97.165497 33.537495 229.502676
std 0.022932 0.028110 9.971957
min -97.203490 33.487960 198.600000
25% -97.182061 33.514003 222.300000
50% -97.165627 33.532457 229.500000
75% -97.148153 33.559099 236.000000
max -97.119257 33.589732 255.600000
From this we can see that all columns have 3774 rows with non-null values. Good! That means we don’t have any NaN or other issues to clean up.
One thing that we do need to fix is the “time” datatype. Pandas is usually pretty good at recognizing datatypes. If it doesn’t, the default assignment is “object”, the same type it assigns to string data. Since this is fundamentally time-series data, we definitely want that to be correctly assigned.
We can use the .astype() function in Pandas to tell it that the data in the “time” column is of type datetime64.
Also, I know that the ride started at about 19:00 CST, but the timestamp on the first trackpoint shows 24:00. The extra +00:00 at the end of each timestamp tells us that the time is in UST, so we’ll also need to correct the records in the “time” column to CST.
The Pandas function .tz_convert() will handle that for us. Here’s a good guide to handling time data in Pandas.
And finally, timezone aware timestamps can cause some issues with calculations, so we’ll want to strip away the timezone indicator with tz.localize().
Aside: In the future it would be good to use the coordinates of the first point to determine time zone and apply the correction automatically.
Also, Pandas seems to intermittently recognize the time data and apply the correct datatype. I left the .astype() example in case yours doesn’t.
# df['time'].astype('datetime64')
df['time'] = df['time'].dt.tz_convert('US/Central')
df['time'] = df['time'].dt.tz_localize(None)
df.info()
df.head(10)
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 3774 entries, 0 to 3773
Data columns (total 4 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 lon 3774 non-null float64
1 lat 3774 non-null float64
2 elev 3774 non-null float64
3 time 3774 non-null datetime64[ns]
dtypes: datetime64[ns](1), float64(3)
memory usage: 118.0 KB
| lon | lat | elev | time | |
|---|---|---|---|---|
| 0 | -97.165271 | 33.488128 | 220.0 | 2017-08-05 18:59:57 |
| 1 | -97.165268 | 33.488129 | 220.0 | 2017-08-05 18:59:59 |
| 2 | -97.165268 | 33.488129 | 220.0 | 2017-08-05 19:00:01 |
| 3 | -97.165269 | 33.488129 | 220.0 | 2017-08-05 19:00:03 |
| 4 | -97.165269 | 33.488130 | 220.0 | 2017-08-05 19:00:05 |
| 5 | -97.165269 | 33.488129 | 220.0 | 2017-08-05 19:00:07 |
| 6 | -97.165268 | 33.488130 | 220.0 | 2017-08-05 19:00:09 |
| 7 | -97.165267 | 33.488127 | 220.0 | 2017-08-05 19:00:11 |
| 8 | -97.165264 | 33.488127 | 220.0 | 2017-08-05 19:00:13 |
| 9 | -97.165261 | 33.488125 | 220.0 | 2017-08-05 19:00:15 |
Basic Visualization
The data is now loaded into a dataframe and basic conditionining and cleanup are finished. Let’s see what it looks like!
We’ll start with some basic plots using plotly.express.
References:
import plotly.express as px
Plotting the Longitude vs Latitude will give a pretty good picture of the track. Since this is a simple XY scatter, it has no notion of coordinate systems, projections, or the ellipsoidal shape of the Earth. Those factors will be important later.
fig_1 = px.scatter(df, x='lon', y='lat', template='plotly_dark')
fig_1.show()
Now let’s look at Elevation vs Time. Since this ride was a loop back to the start, the elevation should end pretty close to where it started. My phone doesn’t have it’s own barometer so the elevation data will have come from RideWithGPS’s digital elevation model. DEM-based elevations should align at the start-finish, so if there’s a disparity, that could point to a problem in the dataset.
fig_2 = px.line(df, x='time', y='elev', template='plotly_dark')
fig_2.show()
Looks like it lines up well.
How about something fancy-ish?
fig_3 = px.scatter_3d(df, x='lon', y='lat', z='elev', color='elev', template='plotly_dark')
fig_3.update_traces(marker=dict(size=2), selector=dict(mode='markers'))
fig_3.show()
THAT is pretty damn cool.
References:
Okay just one more because I ran across it while looking up how to do the plots above.
fig_4 = px.line_mapbox(df, lat='lat', lon='lon', hover_name='time', mapbox_style="open-street-map", zoom=11)
fig_4.update_layout(margin={"r":0,"t":0,"l":0,"b":0})
fig_4.show()
I really cannot express how satisfying that is. A few lines of code and we’ve got GPX track data on a map.
Okay time to get into it for real and deal with distance and speed.
The Earth is not flat
In a simple flat plane, the good ol’ distance formula d = sqrt((x2-x1)^2 + (y2-y1)^2) is all we need to calulate distance, then delta_d/delta_t gives us speed.
Latitude and Longitude represent two points on a spherical surface with respect to a given reference point, or “datum”.
To find the distance between any two points on the surface of a sphere, we must consider that the path is not a straight line but rather follows a circular path across the sphere’s surface. This is known as the “great-circle distance”. Imagine a circle drawn on the surface of the sphere, with the same center, and passing through both points, dividing the sphere into equal halves. This is a “great-circle”. The length of the segment of the circle between the two points is the “great-circle distance”. The Haversine formula is used to calculate the the great-circle distance between two points on a sphere from latitude and longitude.

However, the Earth is not quite spherical. Due to its rotation, the equator bulges out slightly forming an oblate spheroid. This shape is approximated by an ellipsoid. The current standard reference ellipsoid is the World Geodetic System model defined in 1984, known as WGS 84. Calculating the distance between two points on an ellipsoid is significantly more complex than on a sphere. Formulae developed by Vincenty in 1975 handled the problem well, with some exceptions, and were the standard until improved by Karney in 2013.
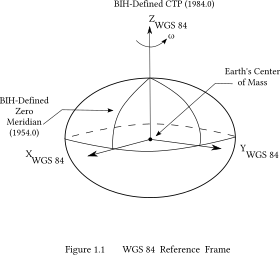
But really the Earth is shaped more like a slightly moldy and battered orange. Because of heterogeneities in the distribution of mass throughout the planet, caused by convection currents in the aesthenosphere and imperfect mixing of minerals. We can map these blobs of higher and lower density by measuring minute variations in the graviational field across the Earth caused by local changes in density.
The latitude and longitude reported by our GPS are geodetic coordinates for points on the WGS 84 reference ellipsoid. The coordinates of the same points on a different reference ellipsoid, North American Datum 1927, for example, would be different, because the NAD 27 reference ellipsoid is a slightly different shape. It’s important to understand this because any algorithm operating on these coordinates must reference the correct ellipsoid model, as does any mapping application.
The geopy package contains methods to calculate the distance using either the spherical method (Haversine) or the geodesic method with your choice of reference ellipsoid (e.g. Vincenty/Karney).
Accounting for elevation change
If we move X miles horizontally on a flat surface, we will have travelled X miles. If that surface were not flat, and we gained some Z miles of elevation, our true distance travelled is the hypotenuse of a triangle with sides X and Z.
The latitude and longitude only give us the location on the surface of a smooth ellipsoid. The GPS track we’re analyzing contains elevation data and, as we saw in the Elevation vs Time plot, it is constantly changing. What is the effect of elevation on the total distance?
Since the distance between points in our GPS track is known to be very small, we can safely ignore the complication of Earth’s curvature and work out the added distance from elevation change with the Euclidean distance formula.
Calculating Distance and speed
To calculate distance and speed we will first need to calculate the distance dist for each segment between a point and the previous point, dist_2d. We will also need the change in elevation, delta_elev, to calculate the contribution of elevation to the distance for dist_3d, and the length of time between recorsd, delta_time, which we’ll use for calculating instantaneous speed.
Out of curiousity, lets run the distance calculations with both spherical and geodesic models. How different will they be at this scale?
from geopy import distance
from math import sqrt, floor
import datetime
# first create lists to store the results, these will be appended ot the dataframe at the end
# Note: i'll be working from the gps_points object directly and then appending results into the dataframe. It would make a lot more sense to operate directly on the dataframe.
delta_elev = [0] # change in elevation between records
delta_time = [0] # time interval between records
delta_sph2d = [0] # segment distance from spherical geometry only
delta_sph3d = [0] # segment distance from spherical geometry, adjusted for elevation
dist_sph2d = [0] # cumulative distance from spherical geometry only
dist_sph3d = [0] # cumulative distance from spherical geometry, adjusted for elevation
delta_geo2d = [0] # segment distance from geodesic method only
delta_geo3d = [0] # segment distance from geodesic method, adjusted for elevation
dist_geo2d = [0] # cumulative distance from geodesic method only
dist_geo3d = [0] # cumulative distance from geodesic method, adjusted for elevation
for idx in range(1, len(gpx_points)): # index will count from 1 to lenght of dataframe, beginning with the second row
start = gpx_points[idx-1]
end = gpx_points[idx]
# elevation
temp_delta_elev = end.elevation - start.elevation
delta_elev.append(temp_delta_elev)
# time
temp_delta_time = (end.time - start.time).total_seconds()
delta_time.append(temp_delta_time)
# distance from spherical model
temp_delta_sph2d = distance.great_circle((start.latitude, start.longitude), (end.latitude, end.longitude)).m
delta_sph2d.append(temp_delta_sph2d)
dist_sph2d.append(dist_sph2d[-1] + temp_delta_sph2d)
temp_delta_sph3d = sqrt(temp_delta_sph2d**2 + temp_delta_elev**2)
delta_sph3d.append(temp_delta_sph3d)
dist_sph3d.append(dist_sph3d[-1] + temp_delta_sph3d)
# distance from geodesic model
temp_delta_geo2d = distance.distance((start.latitude, start.longitude), (end.latitude, end.longitude)).m
delta_geo2d.append(temp_delta_geo2d)
dist_geo2d.append(dist_geo2d[-1] + temp_delta_geo2d)
temp_delta_geo3d = sqrt(temp_delta_geo2d**2 + temp_delta_elev**2)
delta_geo3d.append(temp_delta_geo3d)
dist_geo3d.append(dist_geo3d[-1] + temp_delta_geo3d)
# dump the lists into the dataframe
df['delta_elev'] = delta_elev
df['delta_time'] = delta_time
df['delta_sph2d'] = delta_sph2d
df['delta_sph3d'] = delta_sph3d
df['dist_sph2d'] = dist_sph2d
df['dist_sph3d'] = dist_sph3d
df['delta_geo2d'] = delta_geo2d
df['delta_geo3d'] = delta_geo3d
df['dist_geo2d'] = dist_geo2d
df['dist_geo3d'] = dist_geo3d
# check bulk results
print(f"Spherical Distance 2D: {dist_sph2d[-1]/1000}km \nSpherical Distance 3D: {dist_sph3d[-1]/1000}km \nElevation Correction: {(dist_sph3d[-1]) - (dist_sph2d[-1])} meters \nGeodesic Distance 2D: {dist_geo2d[-1]/1000}km \nGeodesic Distance 3D: {dist_geo3d[-1]/1000}km \nElevation Correction: {(dist_geo3d[-1]) - (dist_geo2d[-1])} meters \nModel Difference: {(dist_geo3d[-1]) - (dist_sph3d[-1])} meters \nTotal Time: {str(datetime.timedelta(seconds=sum(delta_time)))}")
Spherical Distance 2D: 51.73662623269029km
Spherical Distance 3D: 51.74915725516392km
Elevation Correction: 12.53102247363131 meters
Geodesic Distance 2D: 51.740080195107666km
Geodesic Distance 3D: 51.752603641615416km
Elevation Correction: 12.523446507751942 meters
Model Difference: 3.44638645149098 meters
Total Time: 2:17:55
Okay, that is definitely not the most efficient way to do that, but it’ll do for now.
As a quick check, our Total Time is exactly the same as what RWGPS reported. That’s good continued confirmation that the dataset is behaving and doesn’t have anything weird going on.
Did the elevation correction make much of a difference
Not really, but this course also had a maximum elevation difference of around 60 meters. On a course with more topography, I would expect it to be more meaningful, but we can check that another time.
Was there a significant difference between models
Again, not really, just 3 meters over ~52,000 meters travelled. Our points are quite close together relative to the curvature of either model. It is probably resonable to stick with the much simpler great_circle distance. This will be hugely beneficial when running the calculation over 5 years of GPS records.
fig_6 = px.line(df, x='dist_geo3d', y='delta_elev', template='plotly_dark')
fig_6.show()
Here’s a plot of the Elevation Change vs Distance. We saw that the elevation correction was fairly trivial (in this context), just 12 meters over almost 52 kilometers. The greatest elevation difference between two consecutive points was just 2.6 meters. The distance between points would have to be similarly small for that to have a significant effect.
Moving Time
We have Total Activity Time, but what about Moving Time? Most activity analysis apps offer this metric. RideWithGPS says my moving time for this ride was 2:03:05. It is cutting 00:14:50 off the time by dropping records with instantaneous speed below some threshold.
Lets look at a Time vs Distance plot and see if the stationary periods stand out.
fig_5 = px.line(df, x='time', y='dist_geo3d', template='plotly_dark')
fig_5.show()
Yep, every pause is seen as a jog in the Distance vs Time plot (Time continued to increase but Distance did not).
Let’s make a new column with the instantaneous speed in meters per second at each data point and check out a histogram of the results. Then we’ll calculate the total moving time by excluding records below a threshold value. We can run this test for a series of thresholds and see which aligns with RideWithGPS.
df['inst_mps'] = df['delta_geo3d'] / df['delta_time']
fig_8 = px.histogram(df, x='inst_mps', template='plotly_dark')
fig_8.update_traces(xbins=dict(start=0, end=12, size=0.1))
fig_8.show()
I love a Gausian Distribution.
Let’s lop off that stoppage time at the low end and balance it out.
Average human walking speed is about 1.4 meters per second. I’ve had rides that required some walking, so the threshold must be below that.
Note: Python’s range() won’t accept floats, so I brought in numpy to handle it rather than write a list of thresholds manually.
import numpy as np
for threshold in np.arange(0.1, 2.1, 0.1): # delta-distance thresholds from 0 to 2 meters by 0.1 meters
stop_time = sum(df[df['inst_mps'] < threshold]['delta_time'])
print(f"Distance Threshold: {round(threshold,2)}m ==> {str(datetime.timedelta(seconds=stop_time))}")
Distance Threshold: 0.1m ==> 0:12:40
Distance Threshold: 0.2m ==> 0:13:38
Distance Threshold: 0.3m ==> 0:14:04
Distance Threshold: 0.4m ==> 0:14:22
Distance Threshold: 0.5m ==> 0:14:30
Distance Threshold: 0.6m ==> 0:14:34
Distance Threshold: 0.7m ==> 0:14:40
Distance Threshold: 0.8m ==> 0:14:48
Distance Threshold: 0.9m ==> 0:14:50
Distance Threshold: 1.0m ==> 0:15:00
Distance Threshold: 1.1m ==> 0:15:04
Distance Threshold: 1.2m ==> 0:15:04
Distance Threshold: 1.3m ==> 0:15:10
Distance Threshold: 1.4m ==> 0:15:12
Distance Threshold: 1.5m ==> 0:15:18
Distance Threshold: 1.6m ==> 0:15:18
Distance Threshold: 1.7m ==> 0:15:20
Distance Threshold: 1.8m ==> 0:15:20
Distance Threshold: 1.9m ==> 0:15:25
Distance Threshold: 2.0m ==> 0:15:29
It looks like RWGPS is using a threshold of 0.9 m/s instantaneous speed to filter out stoppage time.
Average Speed and Moving Average
Let’s apply the same threshold and calculate Moving Time and Average Moving Speed.
We’ll start by making a new dataframe df_moving with only the records above the threshold. This simplifies the arguments we hand to our functions, but it also takes up more memory. May not be the best way to handle a very large dataset.
df.fillna(0, inplace=True) # fill in the NaN's in the first row of distances and deltas with 0. They were breaking the overall average speed calculation
df_moving = df[df['inst_mps'] >= 0.9] # make a new dataframe filtered to only records where instantaneous speed was greater than 0.9m/s
avg_mps = (sum((df['inst_mps'] * df['delta_time'])) / sum(df['delta_time']))
avg_mov_mps = (sum((df_moving['inst_mps'] * df_moving['delta_time'])) / sum(df_moving['delta_time']))
print(f"Maximum Speed: {round((2.23694 * df['inst_mps'].max(axis=0)), 2)} mph")
print(f"Average Speed: {round((2.23694 * avg_mps), 2)} mph")
print(f"Average Moving Speed: {round((2.23694 * avg_mov_mps), 2)} mph")
print(f"Moving Time: {str(datetime.timedelta(seconds=sum(df_moving['delta_time'])))}")
Maximum Speed: 26.06 mph
Average Speed: 13.99 mph
Average Moving Speed: 15.66 mph
Moving Time: 2:03:05
The average speed over the entire ride was 13.99mph. Filtering down to only “moving” records brings the average speed up to 15.66mph, much better, and it matches with RWGPS’s metrics.
Smoother Plots
When we plot speed vs time, we’re seeing instantaneous speed every 1 to 4 seconds (mostly). That’s a lot of points jumping up and down. The figure isn’t useful to look at. We could down-sample and display fewer points, but that could completely cut out highs and lows. A more elegant way that still preserves the character of the data is a moving average. In this way, we generate a new column that contains the average value of all points a given count before and after. Pandas has a function called .rolling() that does exactly that.
df['avg_mps_roll'] = df['inst_mps'].rolling(20, center=True).mean()
fig_16 = px.line(df, x='time', y=['inst_mps', 'avg_mps_roll'], template='plotly_dark') # as of 2020-05-26 Plotly 4.8 you can pass a list of columns to either x or y and plotly will figure it out
fig_16.show()
Elevation Gain
Another metric we still need is the total elevation gain. The author of the original article wrote a custom function to return only positive inputs, then used map to apply it to df['delta_elev'] and generate a temporary list of positive values.
def positive_only(x):
if x > 0:
return x
else:
return 0
pos_only = list(map(positive_only, df['delta_elev']))
print(f"Total Elevation Gain: {round(sum(pos_only), 2)}")
Total Elevation Gain: 381.2
That’s fine but I think it can be simplified, or at least compressed with lambda and filter.
elev_gain = sum(list(filter(lambda x : x > 0, df['delta_elev'])))
print(f"Total Elevation Gain: {round(elev_gain, 2)}")
Total Elevation Gain: 381.2
filter is like map but as the name implies, it is a filter on the series you apply it to. If the function evaluates True, it passes the value from the series, if not, the value is skipped.
References:
But is there a pure-Pandas way to do it? Can we simplify further?
print(f"Elevation Gain: {round(sum(df[df['delta_elev'] > 0]['delta_elev']), 2)}")
print(f"Elevation Loss: {round(sum(df[df['delta_elev'] < 0]['delta_elev']), 2)}")
Elevation Gain: 381.2
Elevation Loss: -381.4
There is, actually, he used it earlier and it’s pretty slick. It’s called boolean indexing. df['delta_elev'] > 0 returns the row indices for every value in delta_elev that is positive. We used those indices in the next level up to select the values we want to sum, e.g. df[idx]['delta_elev'] and sum them together. I wrapped it in round() to clean up the output.
References:
Wrapping Up
We’ve been able to replicate most of the metrics that sites like Strava, Garmin, and RideWithGPS present for activities.
- Distance
- Elevation Gain/Loss
- Total Time
- Stopped Time
- Moving Time
- Max Speed
- Average Speed
- Moving Average Speed
Average Pace and Moving pace are transformatons of Average Speed and Moving Speed.
Max Grade and Average Grade are simple calculations on delta_elev and segment length, as are Ascent Time and Descent Time.
VAM and Calories will take some further research to understand.
Next Steps
In the long term, we’ll analyze a large number of .gpx files. There’s a tongue-in-cheek axiom in cycling that goes “It doesn’t get easier, you just go faster.” Seems like an easy one to test since I’ve got 5 years of cycling records to work with.
The challenge will be handling that volume of points efficiently. At the least, I don’t want to read-in hundreds of .gpx over and over while working out the code. Some kind of external storage like JSON or CSV would be a simple solution, but an external database like PostGIS (built on PostgreSQL) might be better, and I’d learn some database handling finally. As an added bonus, it can be read by QGIS.
Once all the .gpx are usefully gathered and today’s code has been collapsed and made more efficient, we’ll take a crack at the ever satisfying heatmap, probably with folio and Mapbox.
